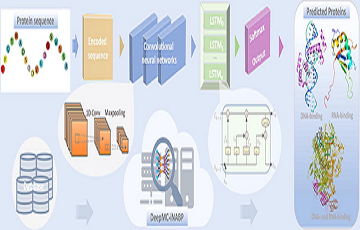
DeepMC_iNABP
Deep learning for multiclass identification and classification of nucleic acid-binding proteins.
Go to service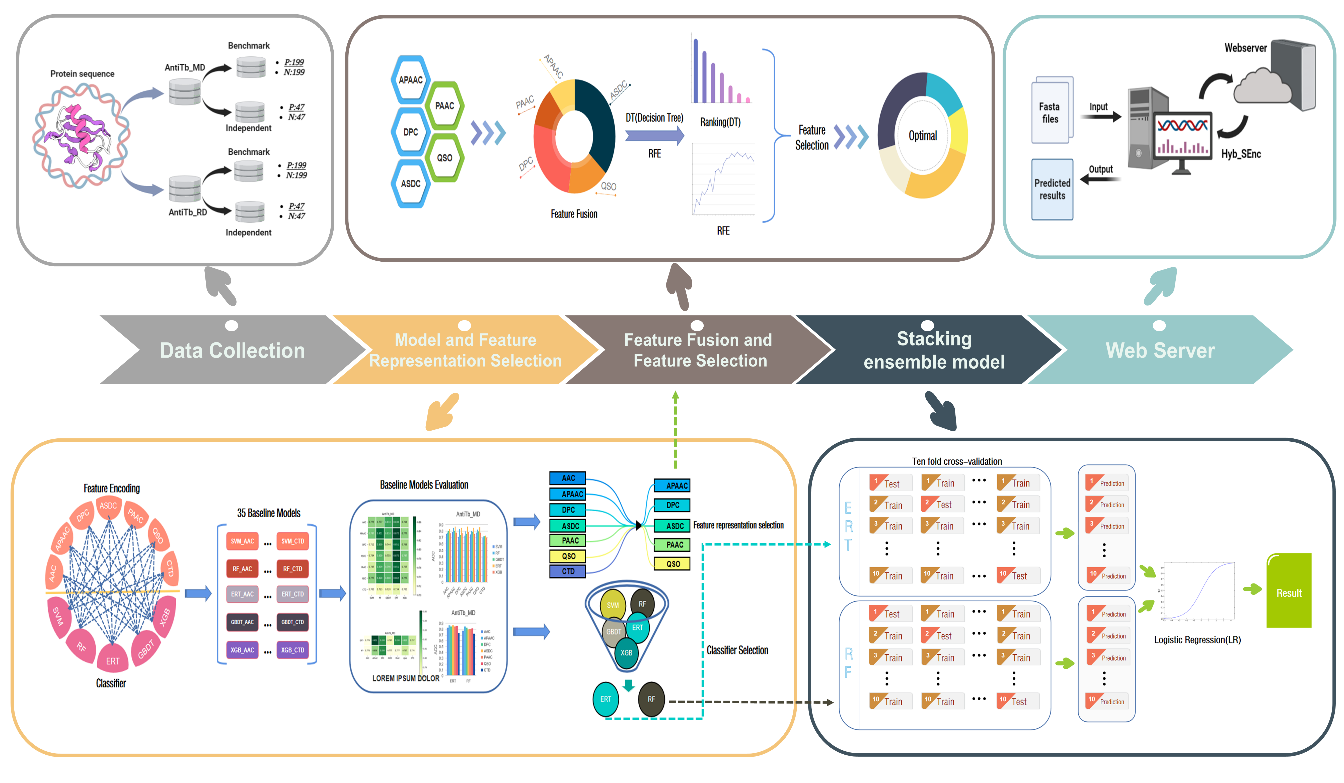
Hyb_SEnc
An Antituberculosis Peptide Predictor Based on a Hybrid Feature Vector and Stacked Ensemble Learning.
Go to service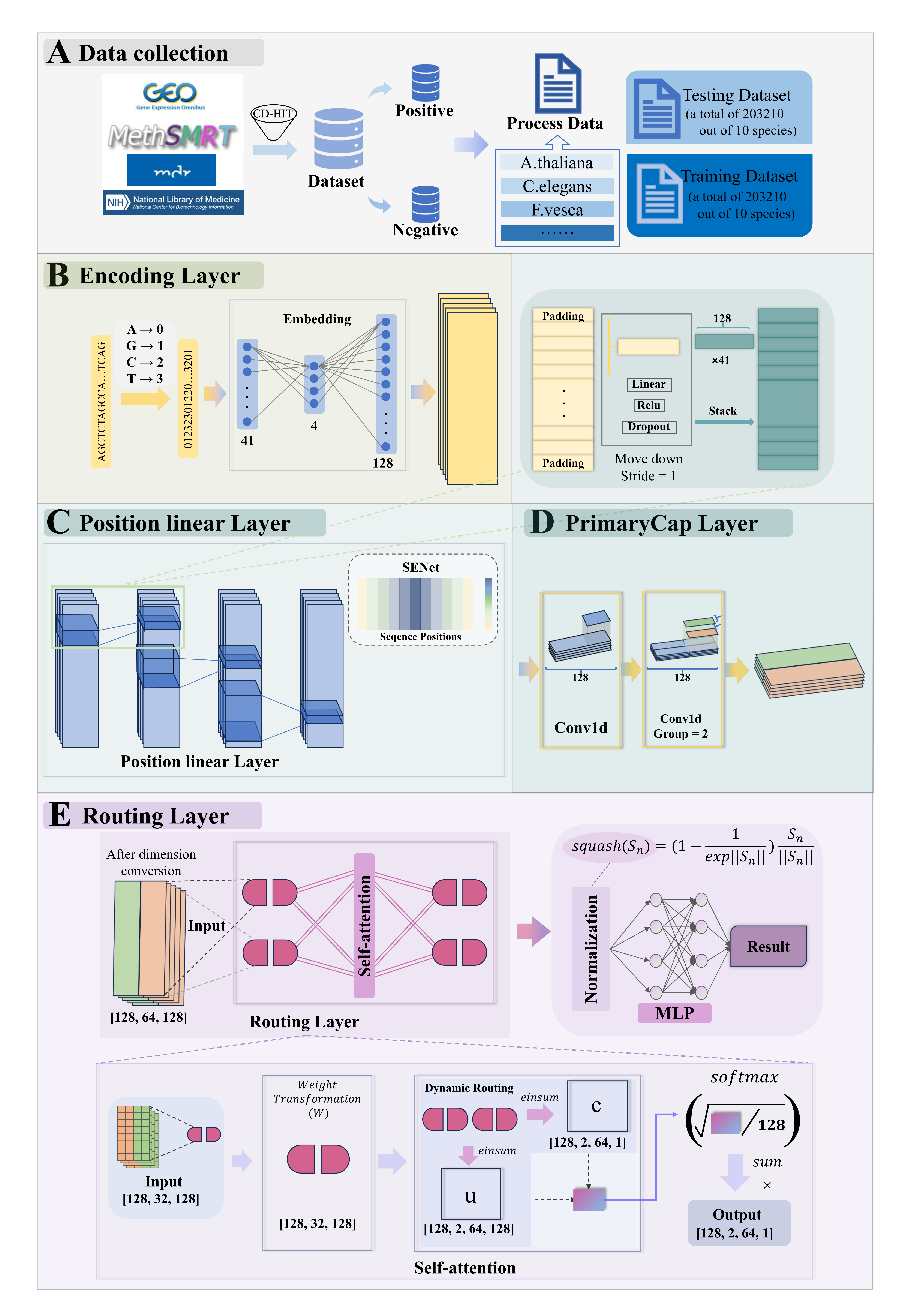
PSAC_6mA
6mA site identifier using self-attention capsule network based on sequence-positioning.
Go to service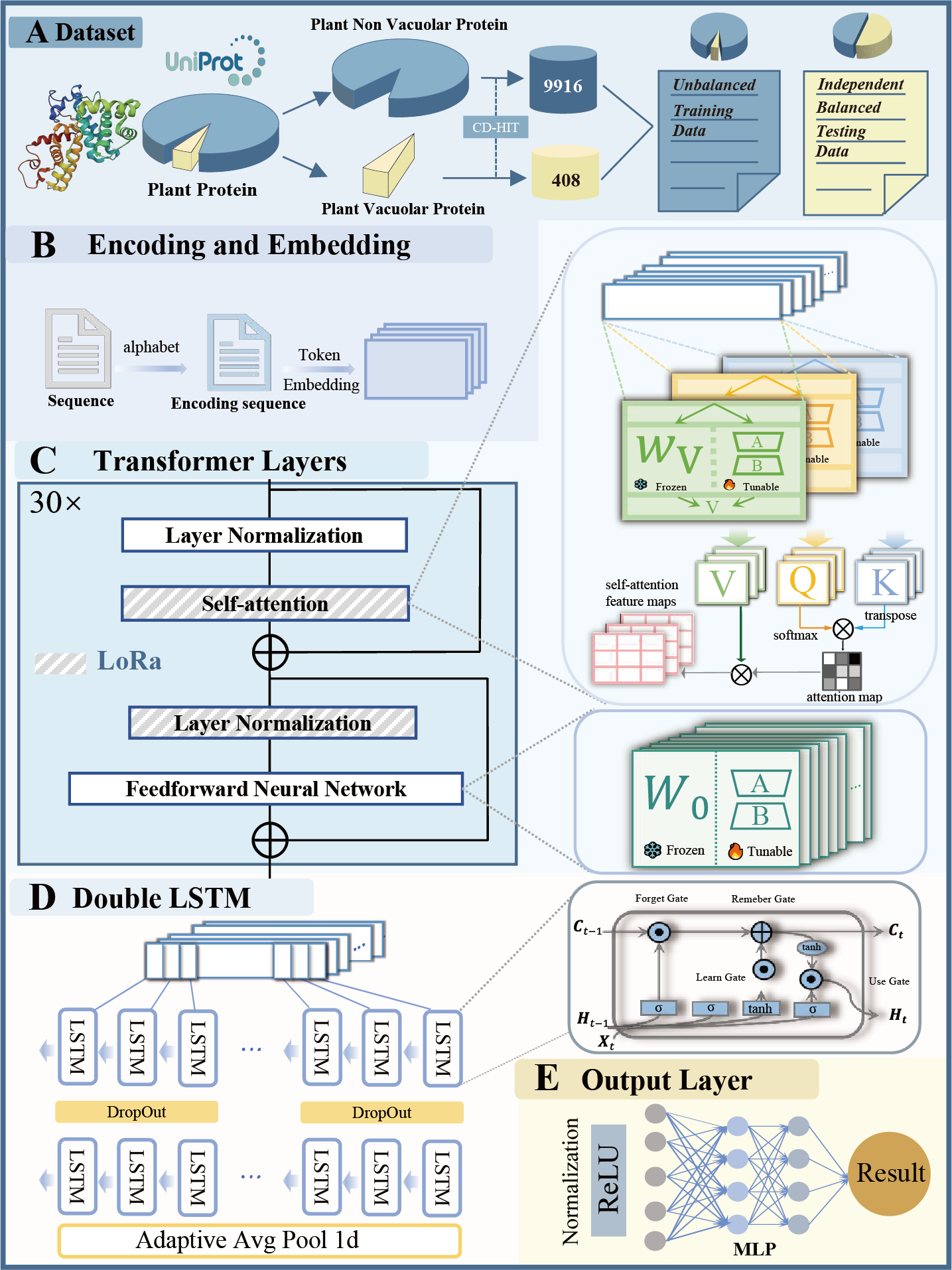
PEL_PVP
Application of Plant Vacuolar Protein Identifier Based on Low-rank Fine-tuning of ESM2 Model Using LoRa Technology and bilayer LSTM on Unbalanced Datasets
Go to service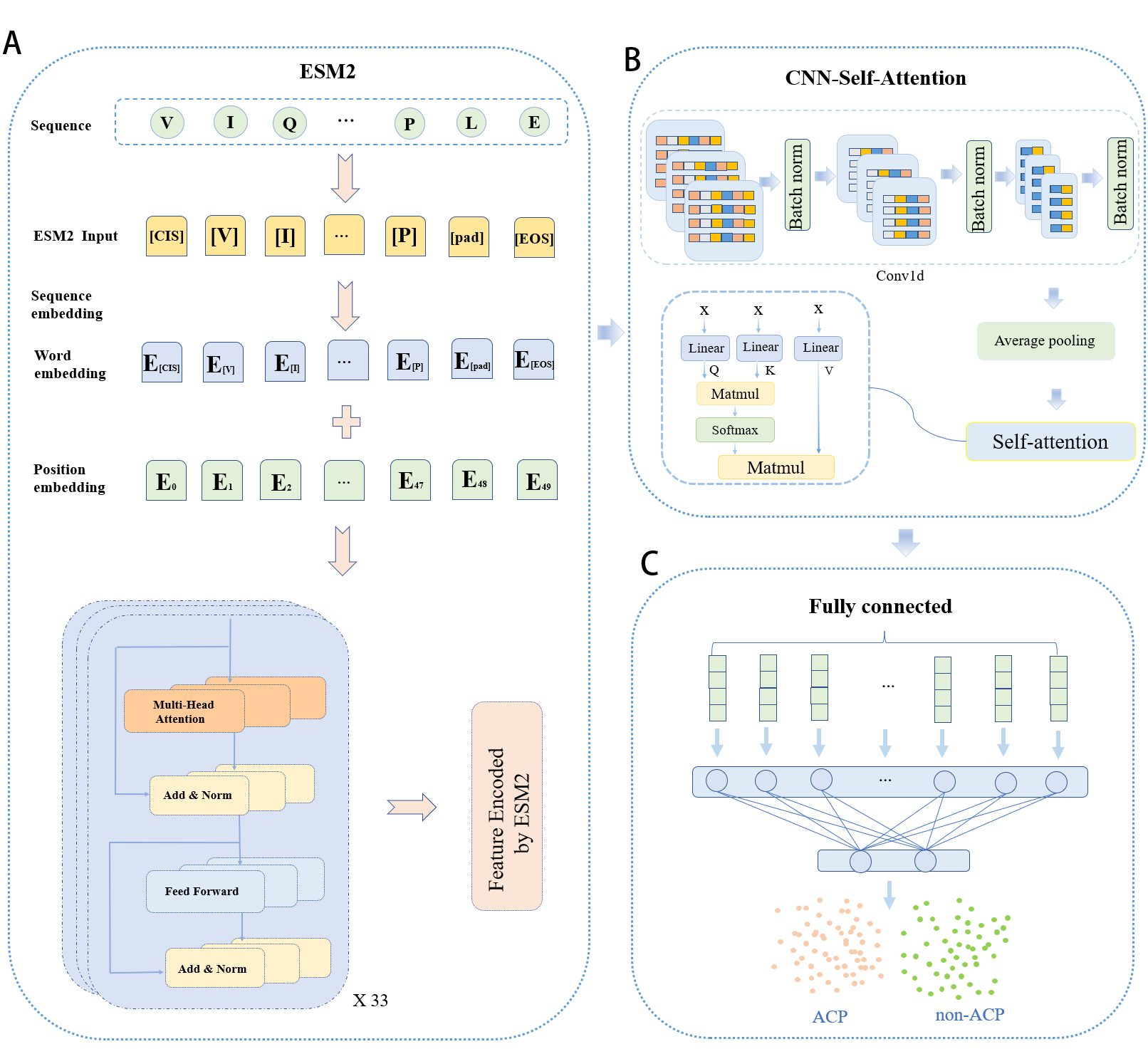
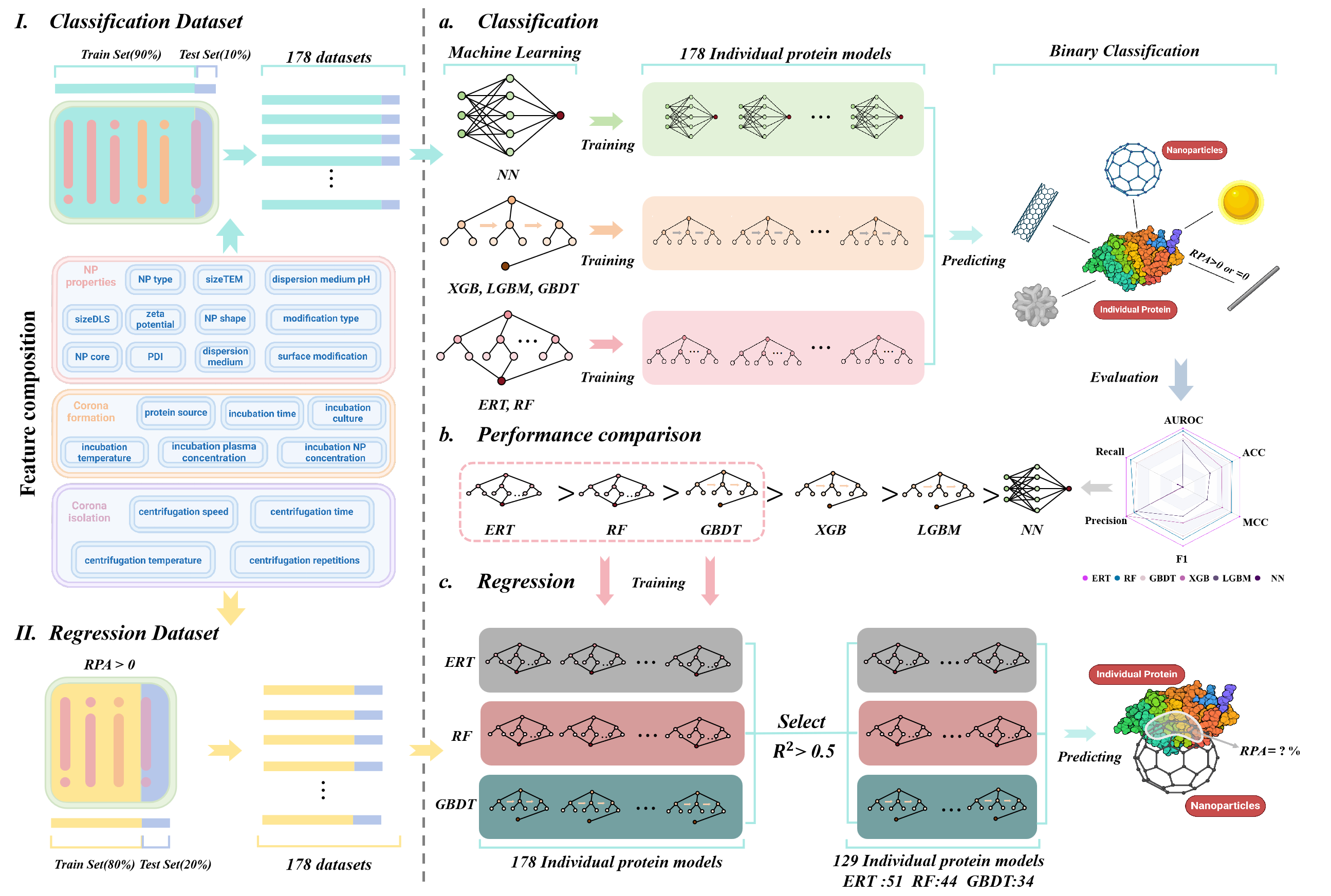
PC_ML
Machine learning enables comprehensive prediction of the relative protein abundance of multiple proteins on the protein corona.
Go to service
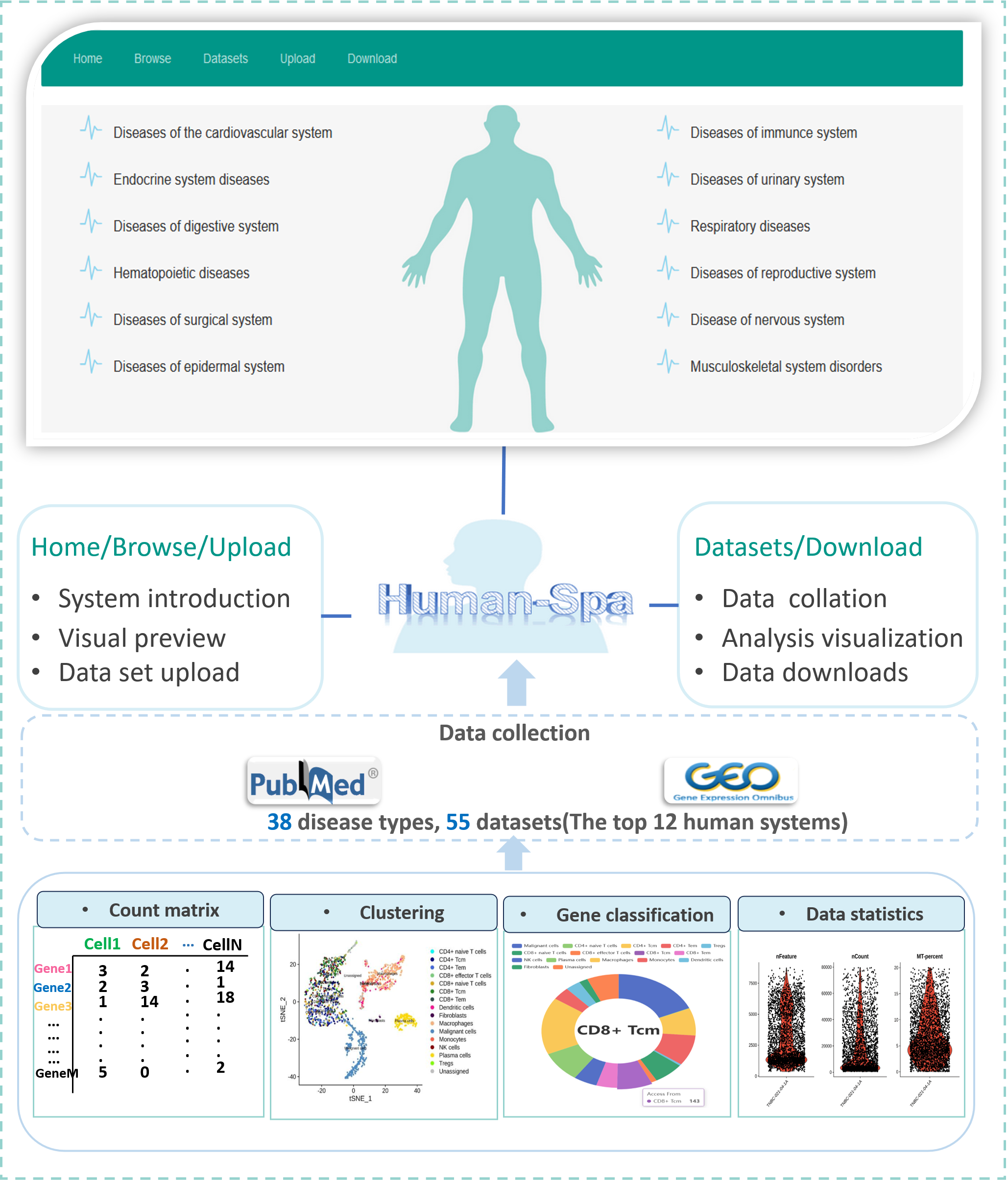
Human-Spa
An Online Platform Based on Spatial Transcriptome Data for Diseases of Human Systems.
Go to service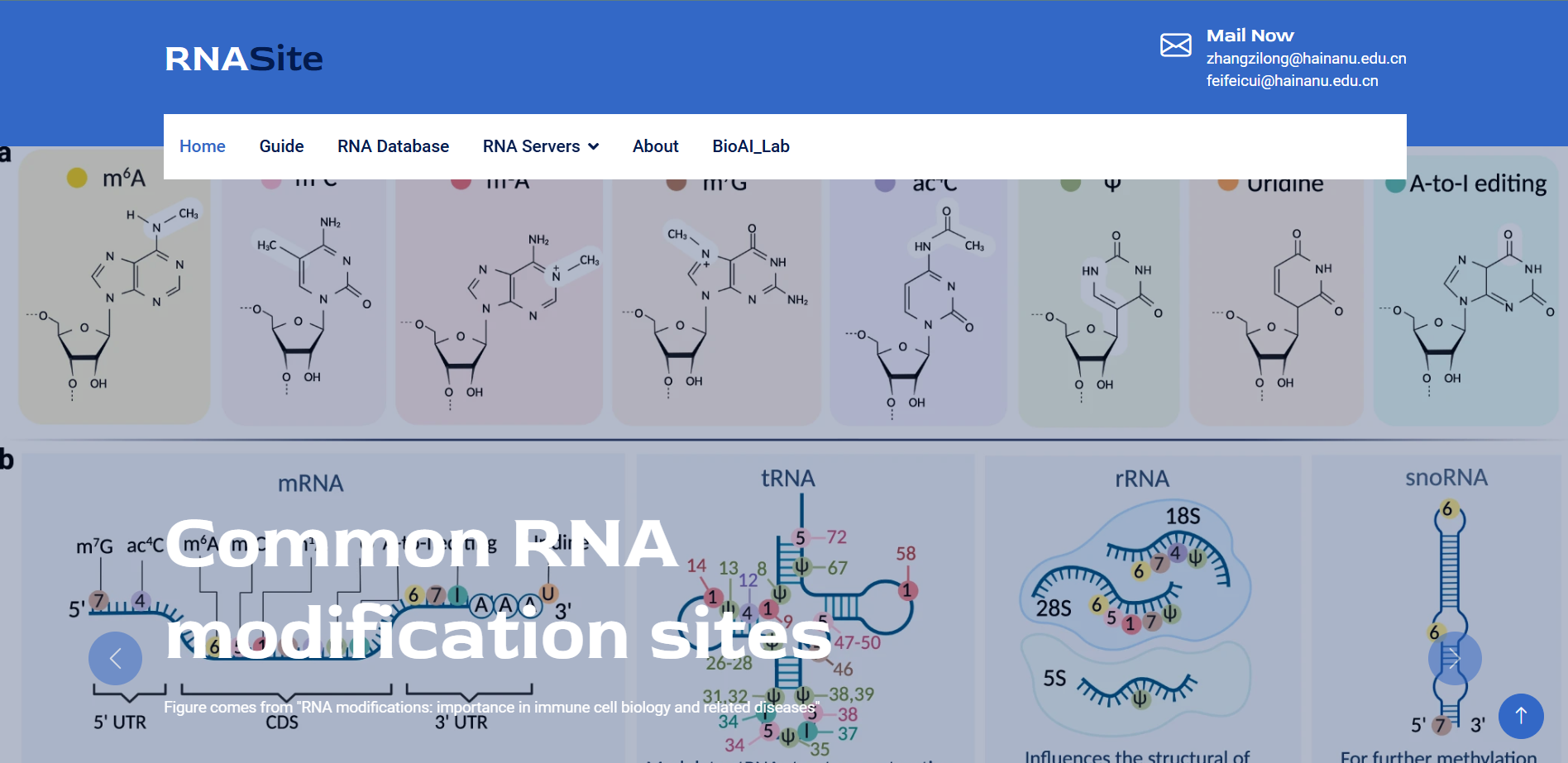
RNA Site
A one-stop tool website that integrates multiple RNA site databases and servers.
Go to service
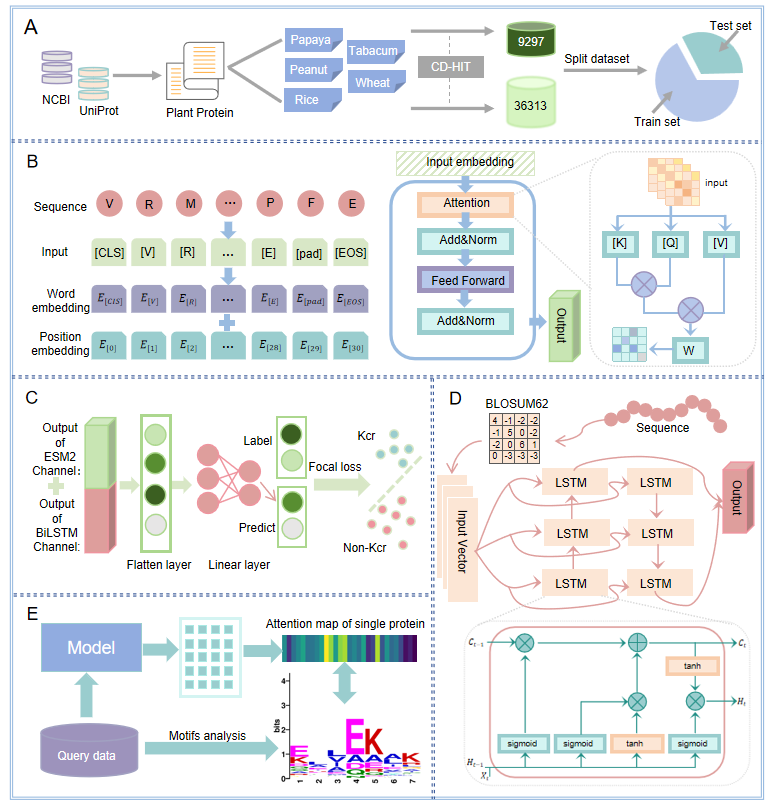
DeepNhKcr
explainable deep learning framework for the prediction of crotonylation sites of non-histone lysine in plants based on pre-trained protein language model.
Go to service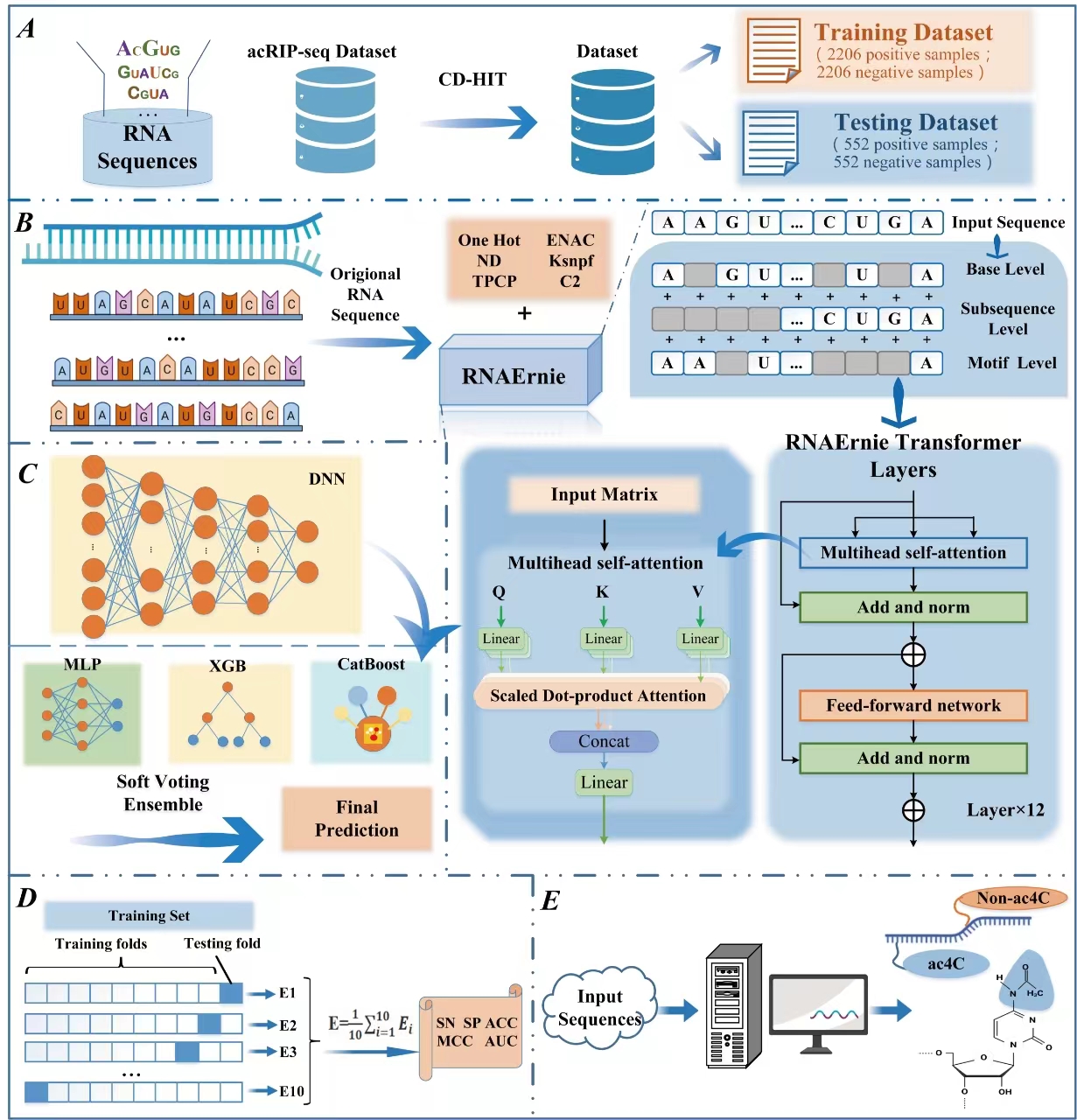
Voting-ac4C
Pre-trained large RNA language model enhances RNA N4-acetylcytidine site prediction.
Go to service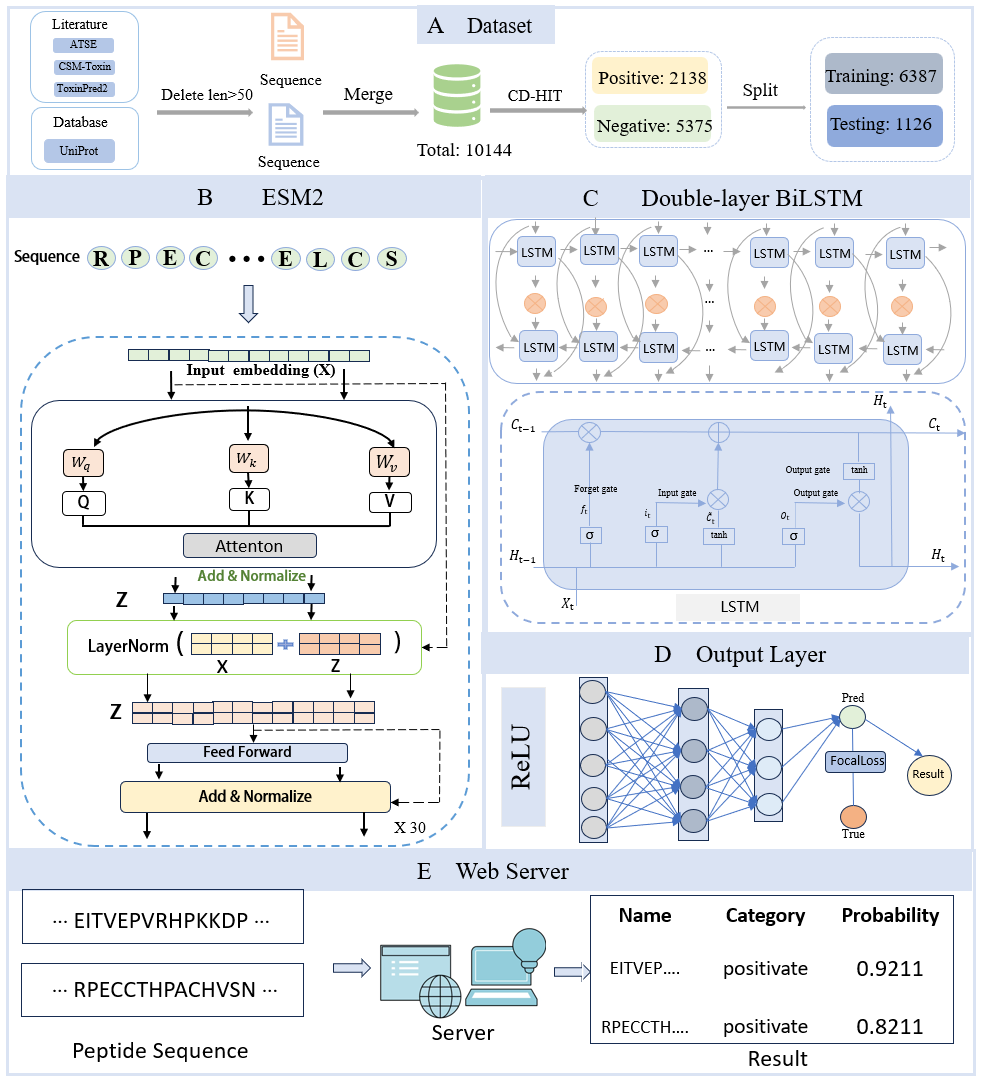
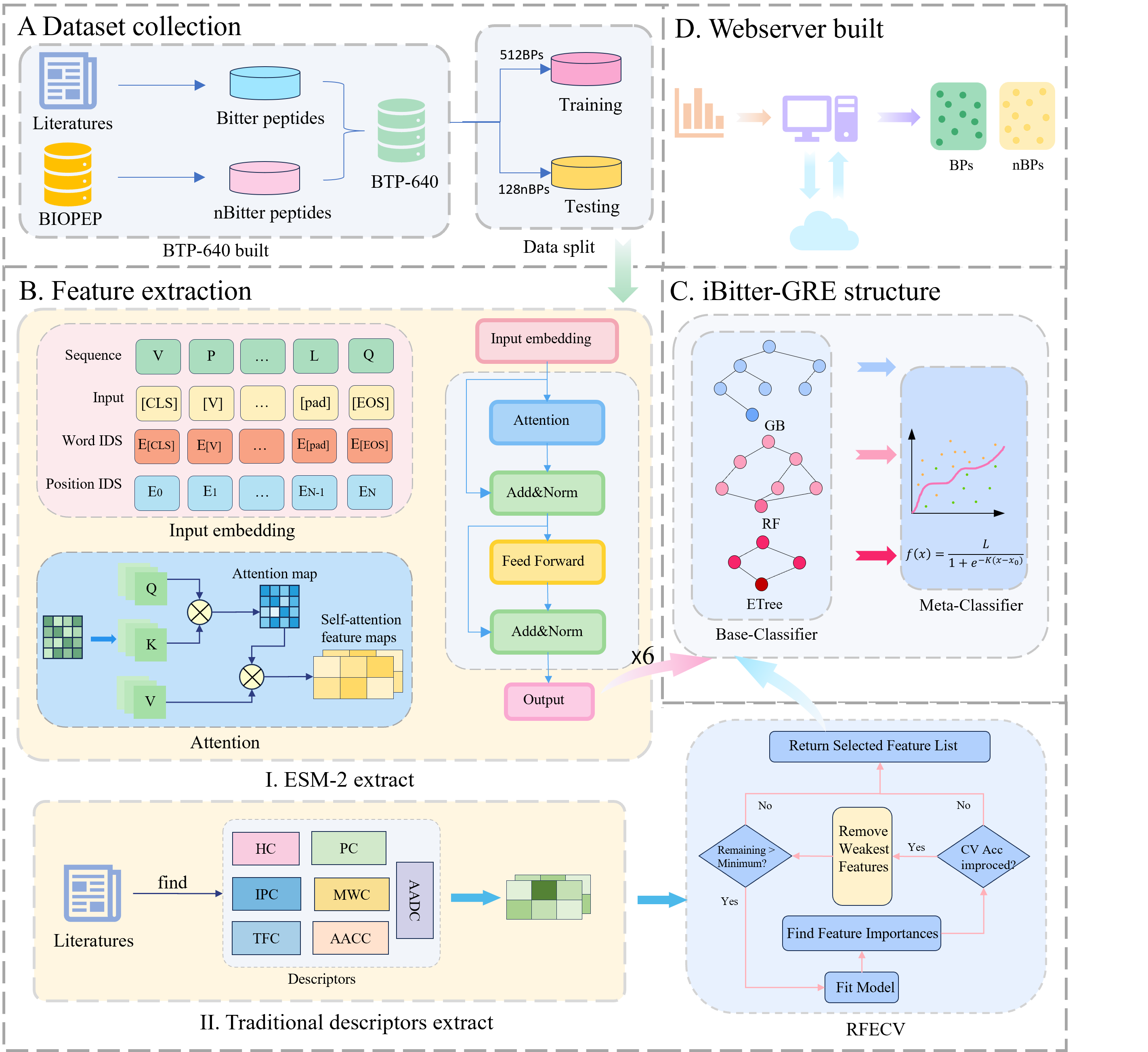
iBitter-GRE
A Novel Stacked Bitter Peptide Predictor with ESM-2 and Multi-View Features.
Go to service
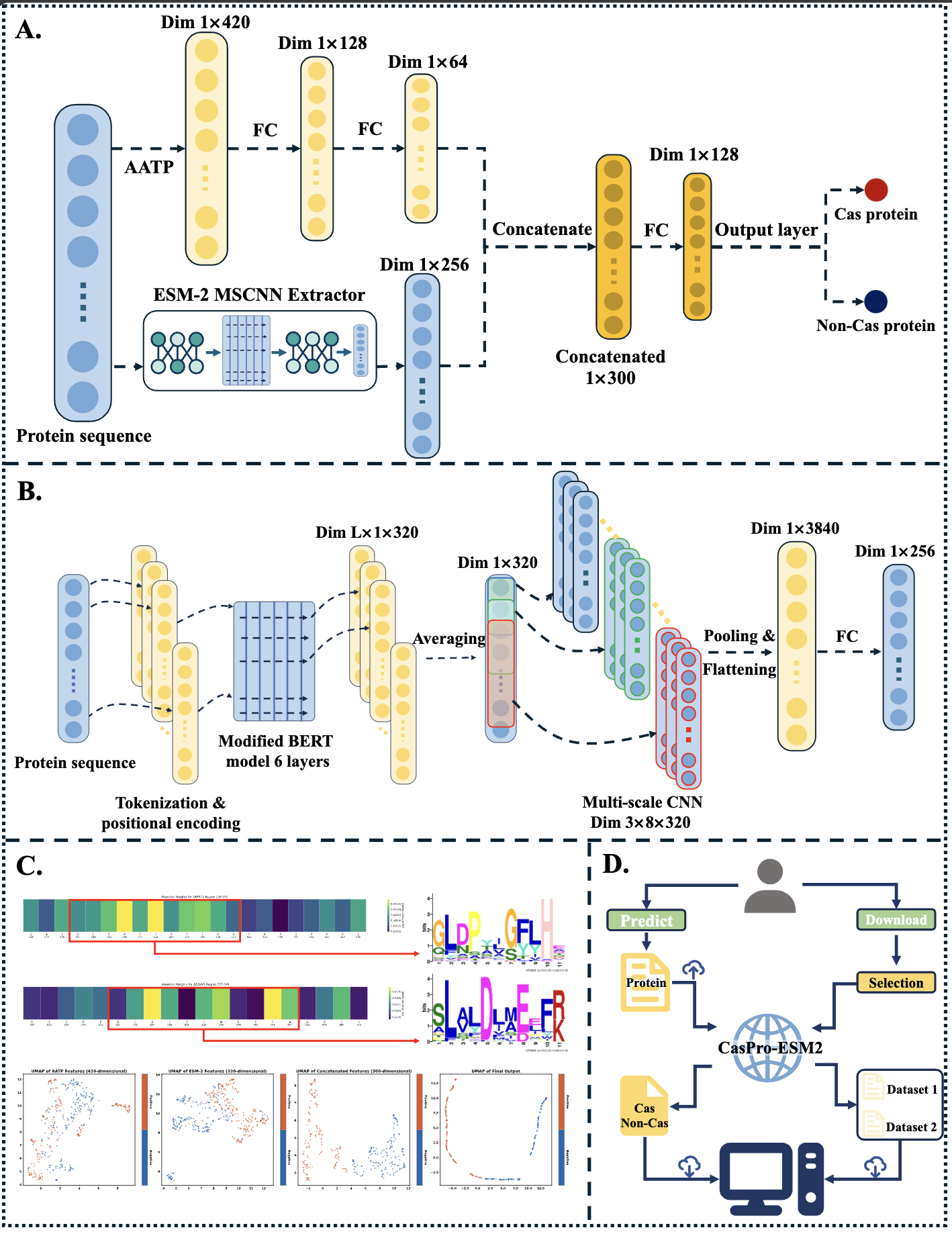
CasPro-ESM2
A Tool for Identifying Cas Proteins Based on the ESM-2 Protein Language Model.
Go to service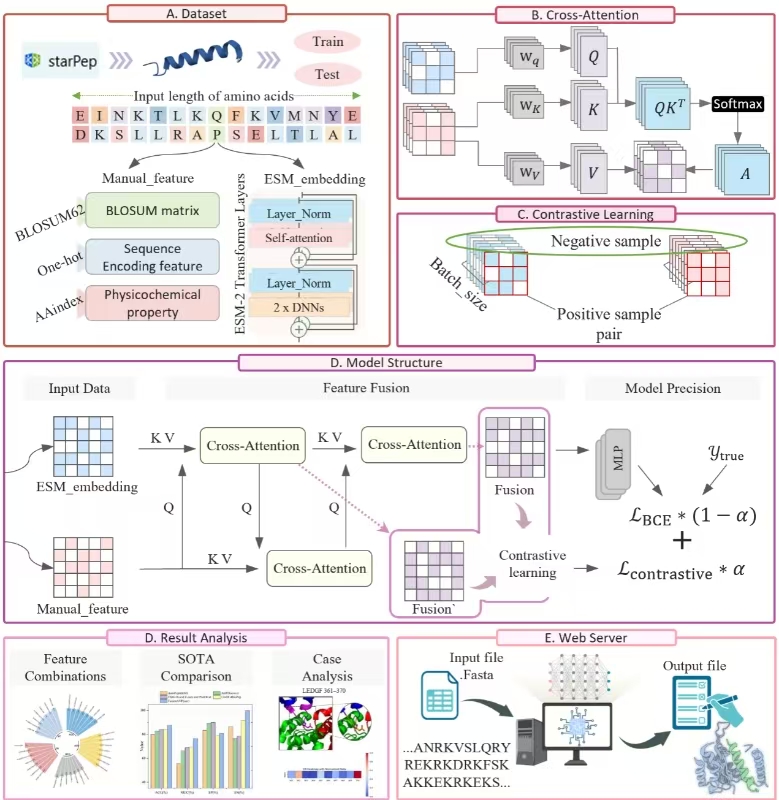
FusionAVP
Multimodal Fusion Strategy for Enhancing Robust Antiviral Peptide Classification.
Go to service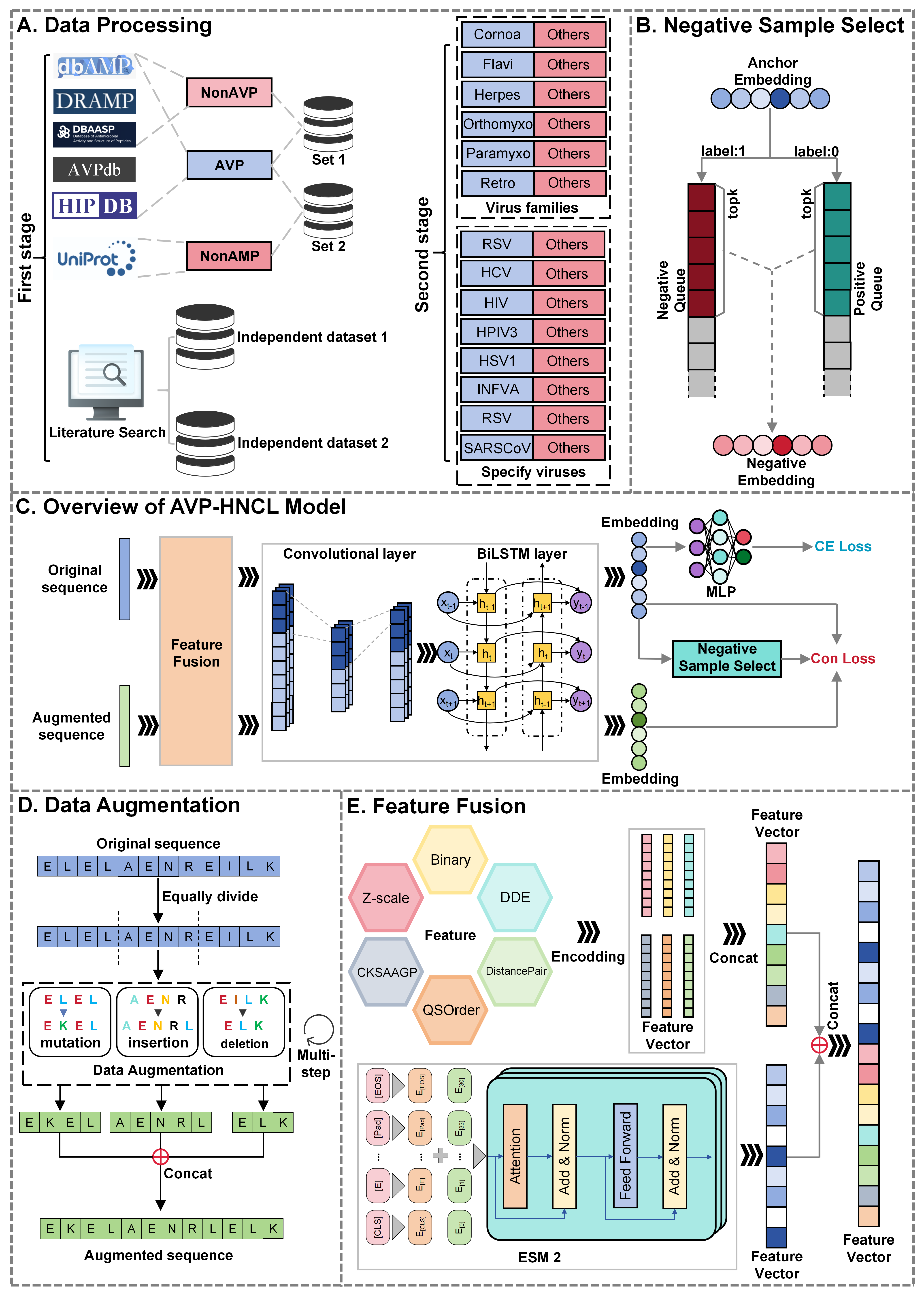
AVP-HNCL
Innovative Contrastive Learning with Queue-based Negative Sampling Strategy for Dual-Phase Antiviral Peptide Prediction.
Go to service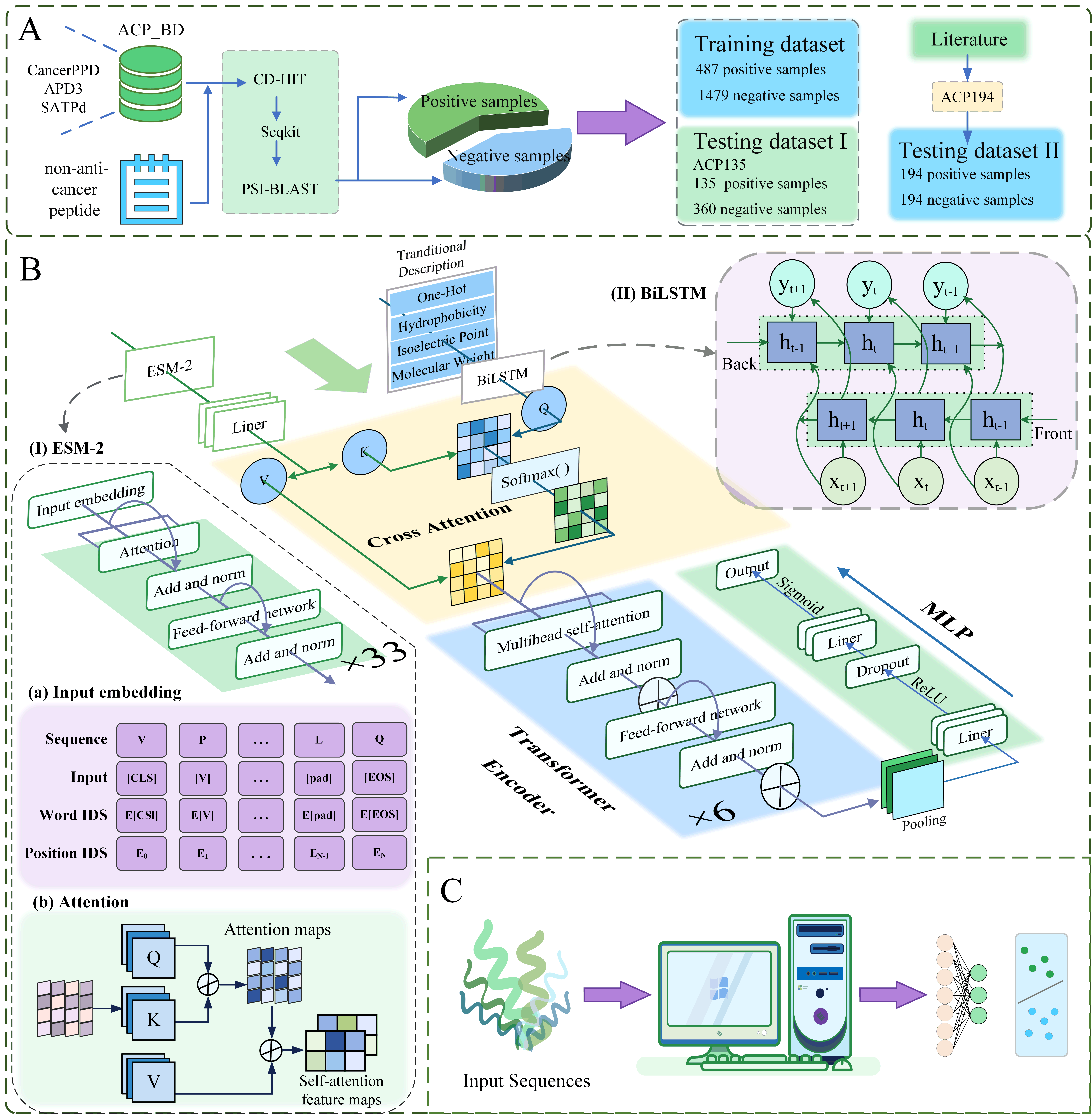
ACP-EPC
An Interpretable Deep Learning Framework for Anticancer Peptide Prediction Utilizing Pre-trained Protein Language Model and Multi-view Feature Extracting Strategy.
Go to service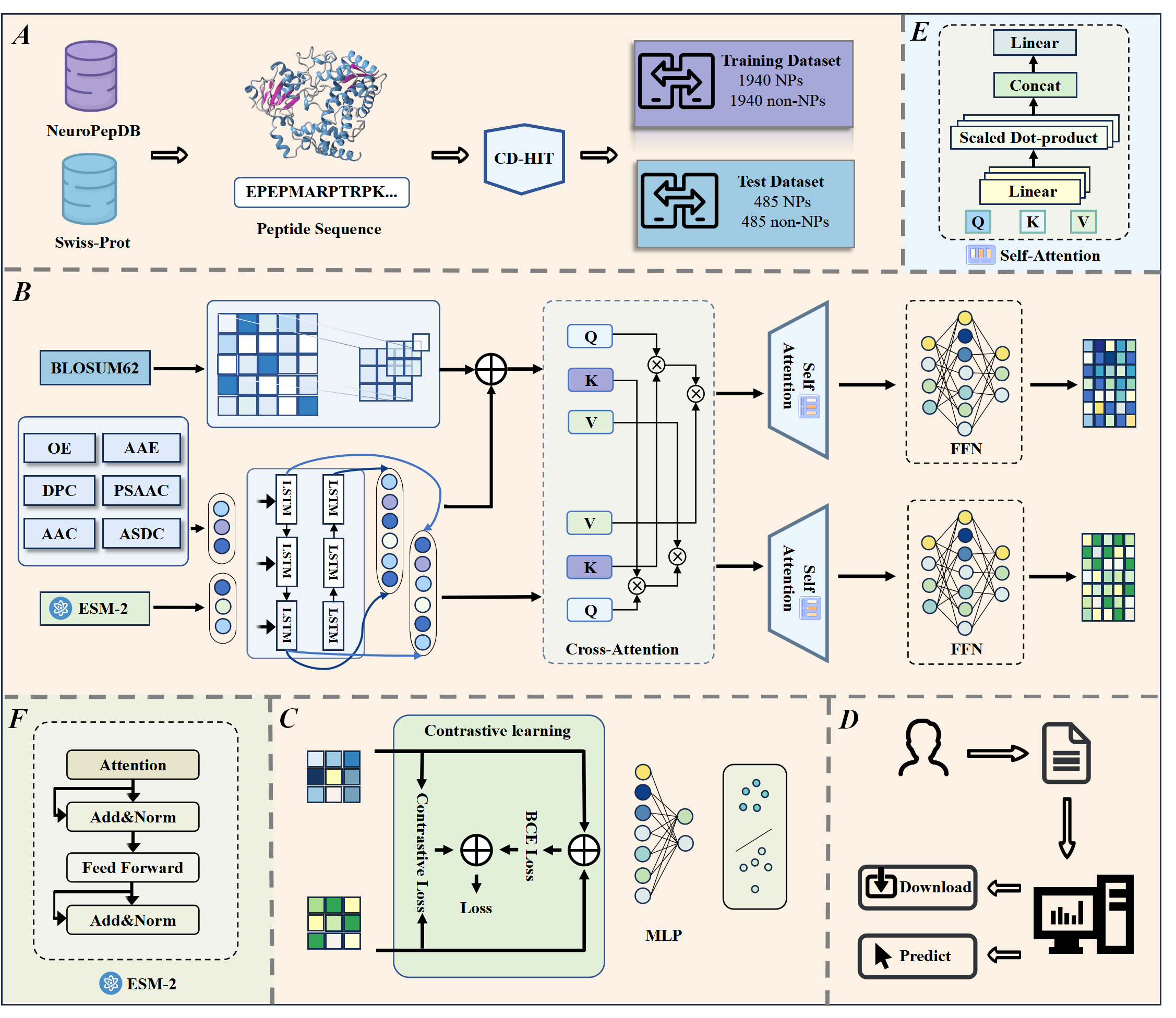
NeuroCL
A deep-learning approach for identifying neuropeptides based on contrastive learning
Go to service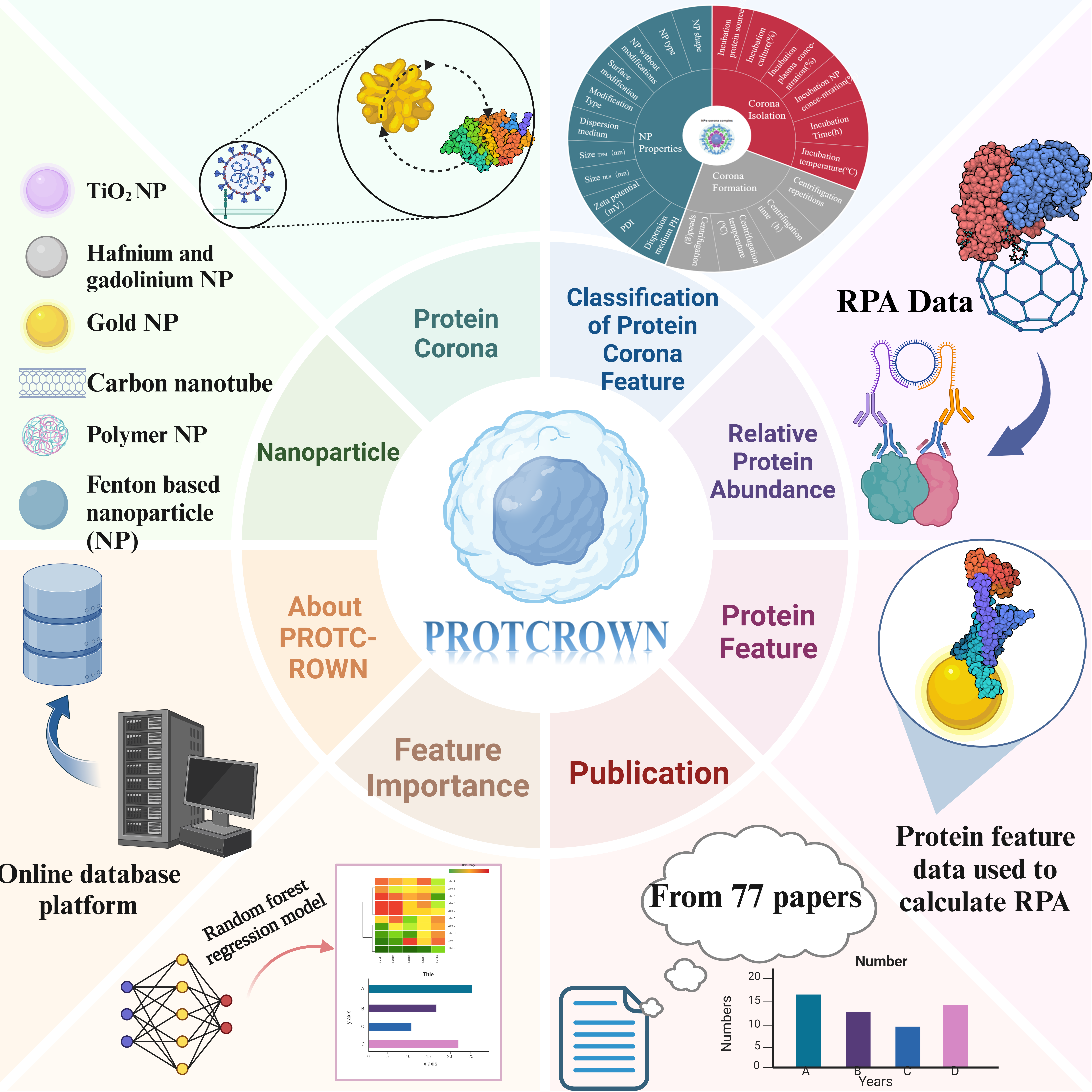
PROTCROWN
A Manually Curated Resource of Protein Corona Data for Unlocking the Potential of Protein–Nanoparticle Interactions.
Go to service
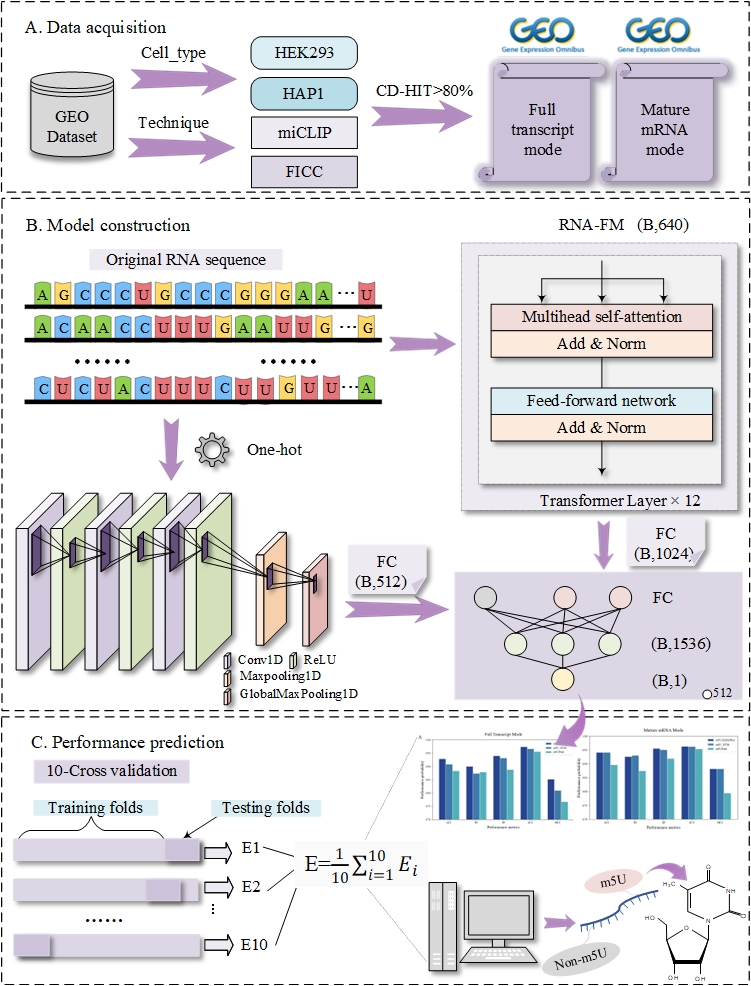
m5U-HybridNet
Integrating an RNA Foundation Model with CNN Features for Accurate Prediction of 5-Methyluridine Modification Sites.
Go to service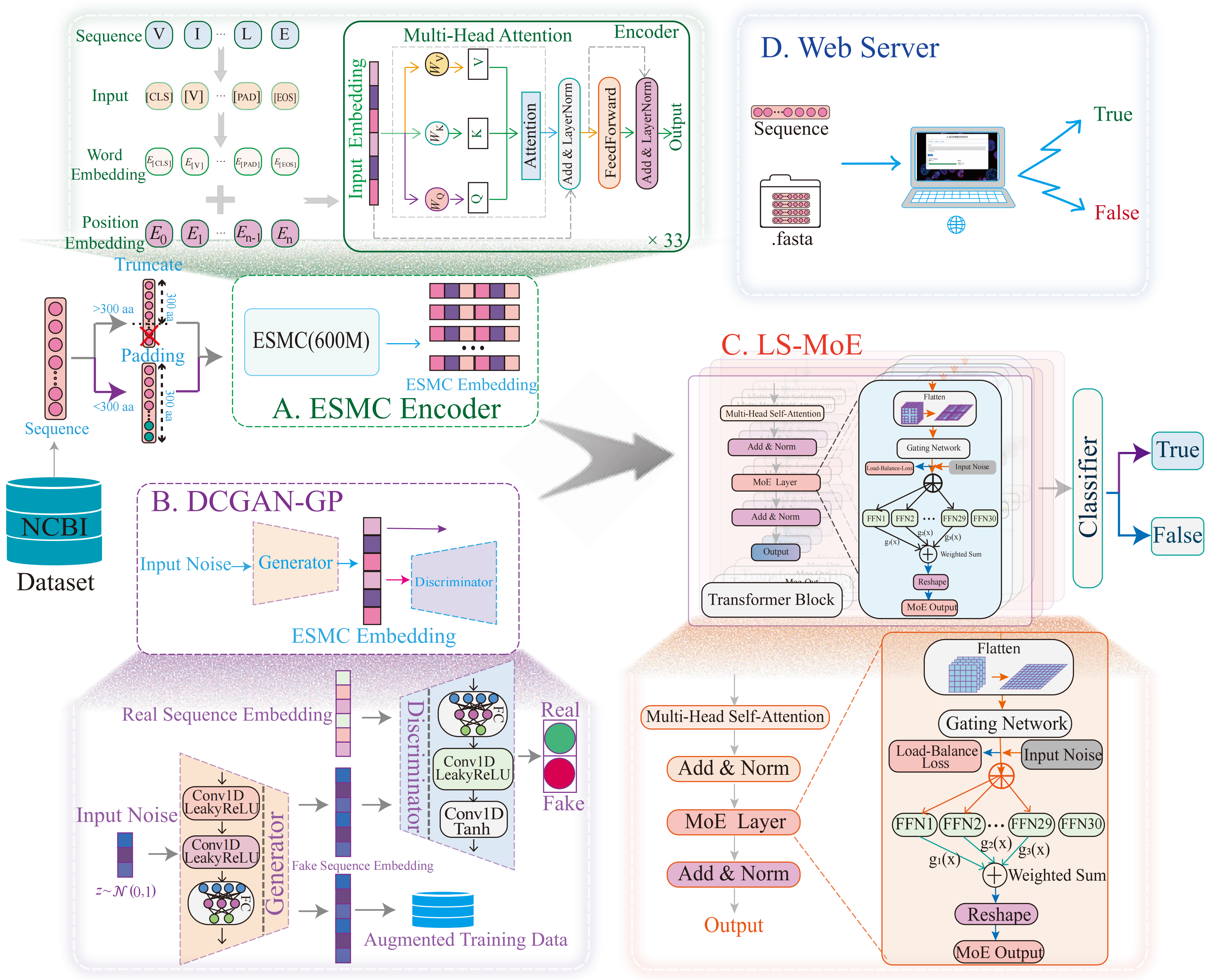
AcidNetPro
Acidophilic Protein Classification via DCGAN-GP Enhanced Embeddings and Lightweight Sparse MoE Transformer.
Go to service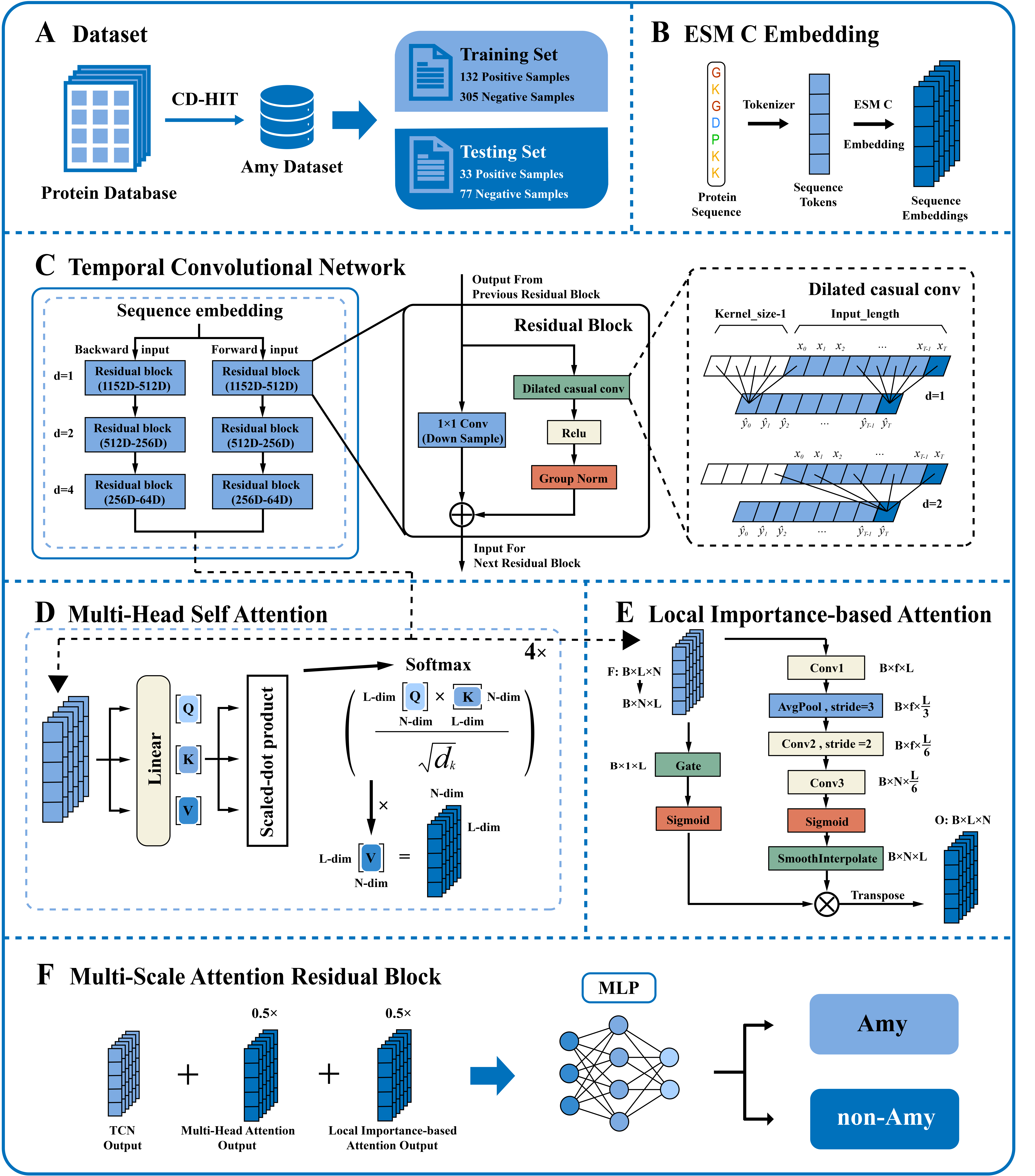
FA-Amy
An Amyloid Protein Prediction Model Based on Protein Pre-trained Large Models and an Attention-Fusion Strategy.
Go to service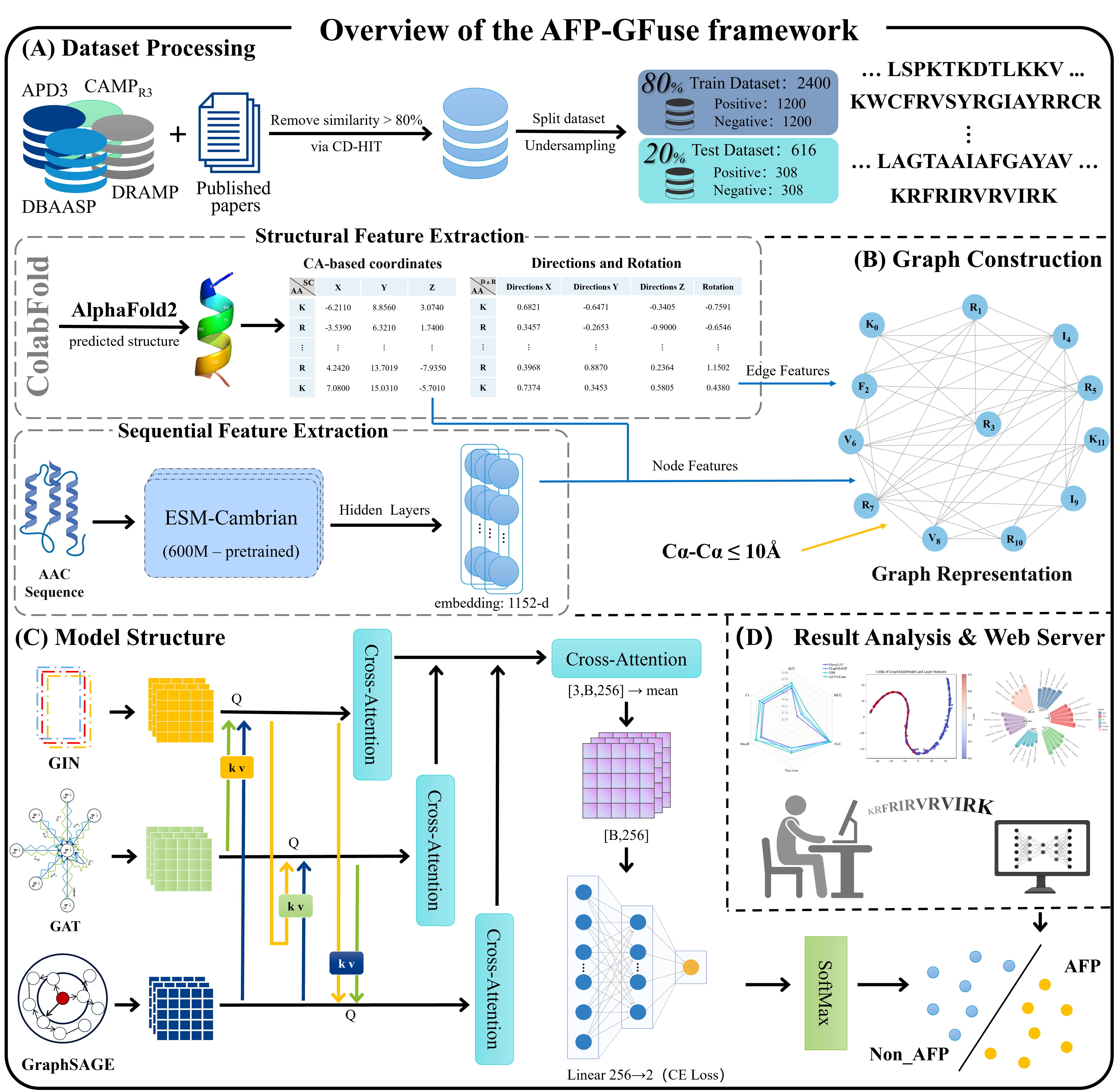
AFP-GFuse
An Antifungal Peptide Identification Model with Structural Information Fusion via Multi-Graph Neural Networks and Cross-Attention Mechanism
Go to service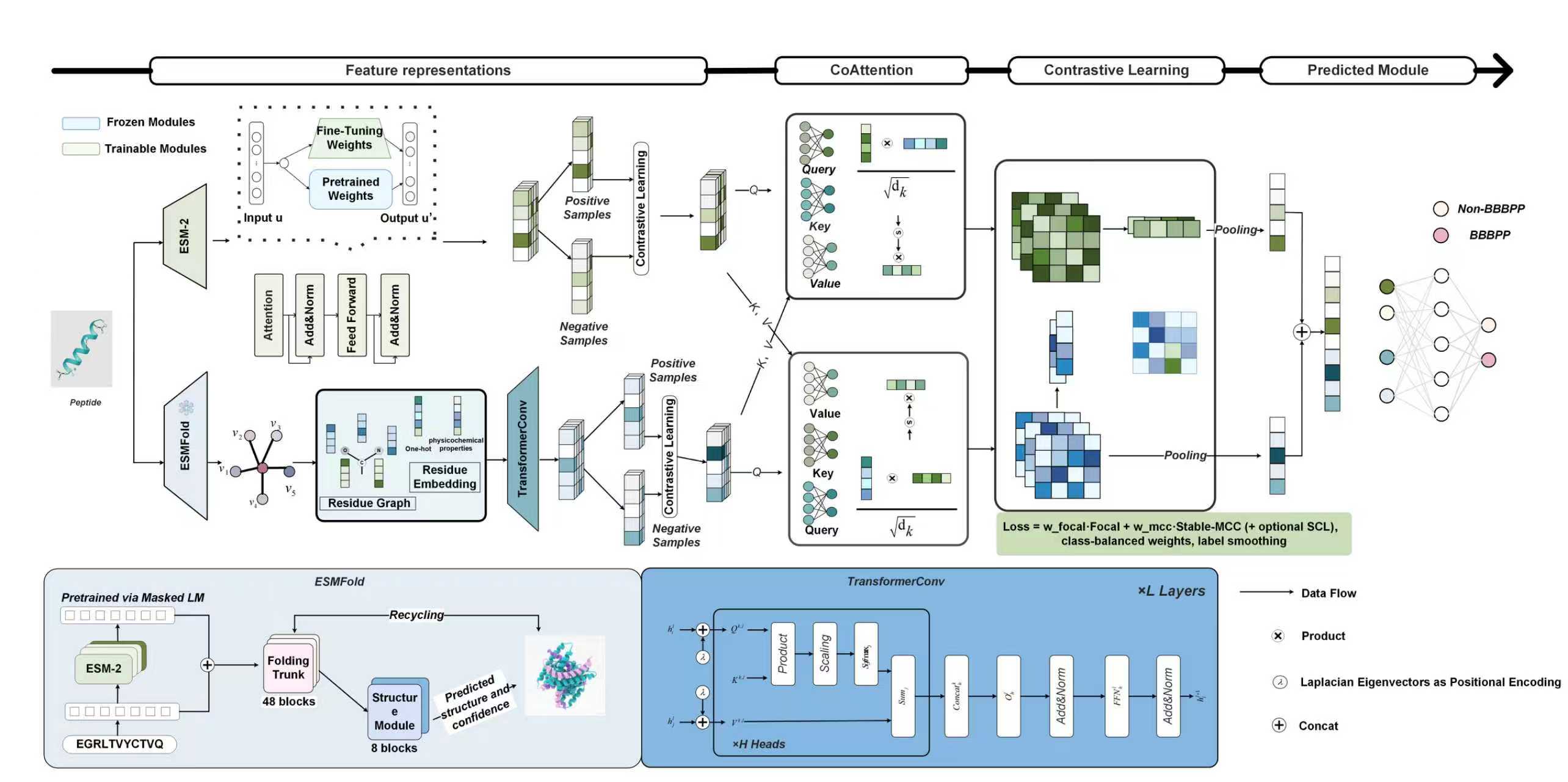
INB³P
A Multi-Modal and Interpretable Co-Attention Framework Integrating Property-Aware Explanations and Memory-Bank Contrastive Fusion for Blood–Brain Barrier Penetrating Peptide Discovery
Go to service